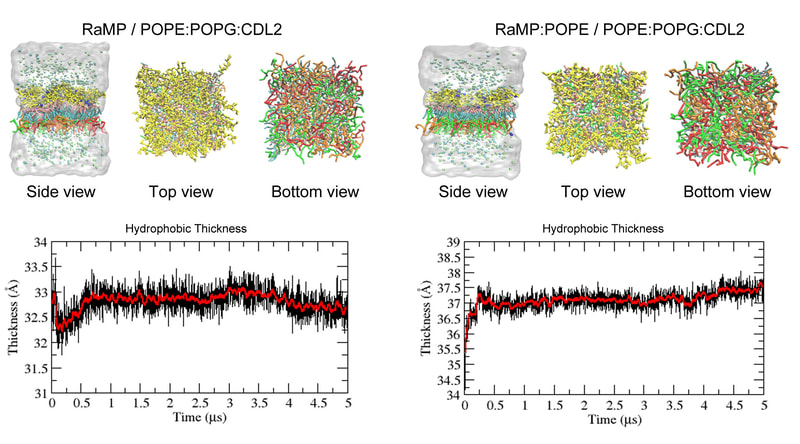
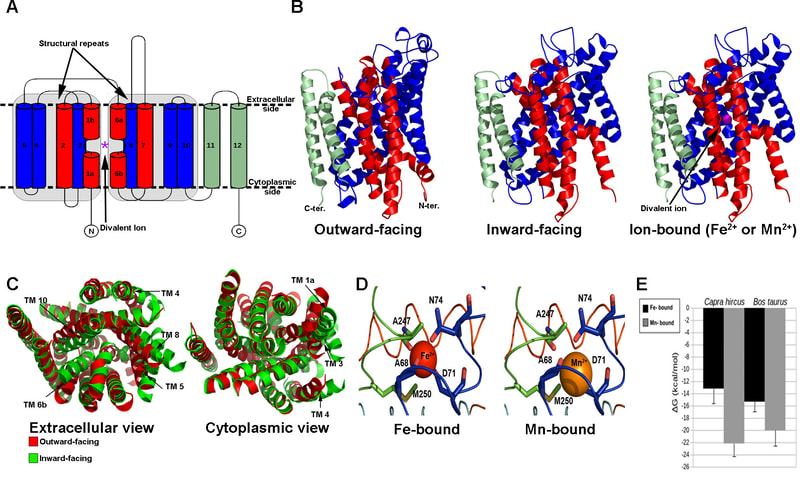
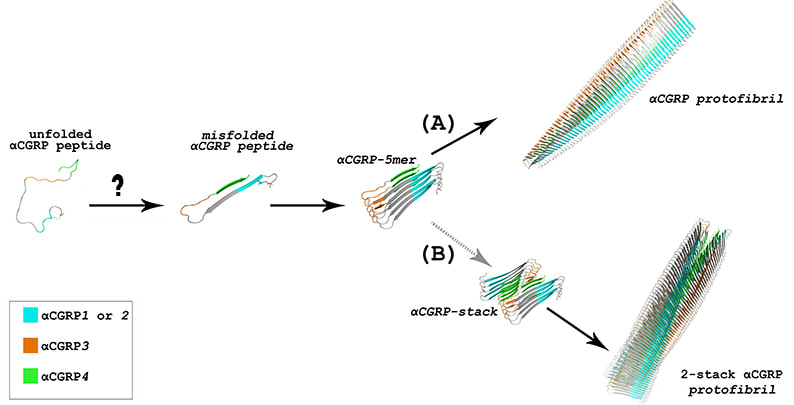
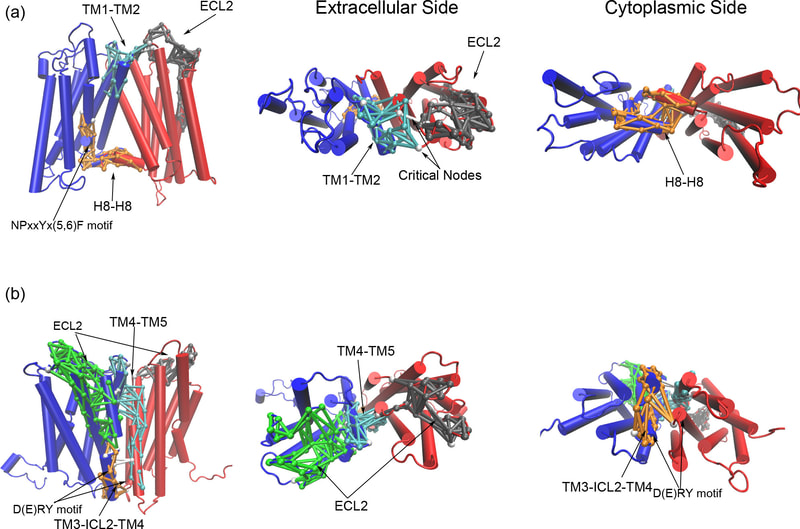
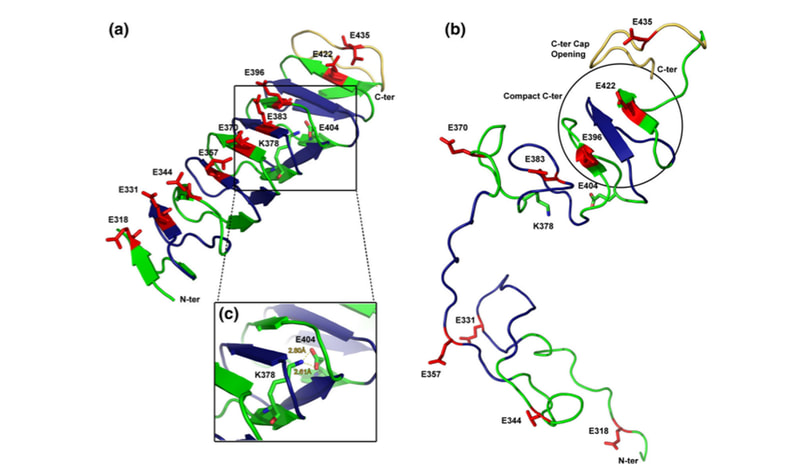
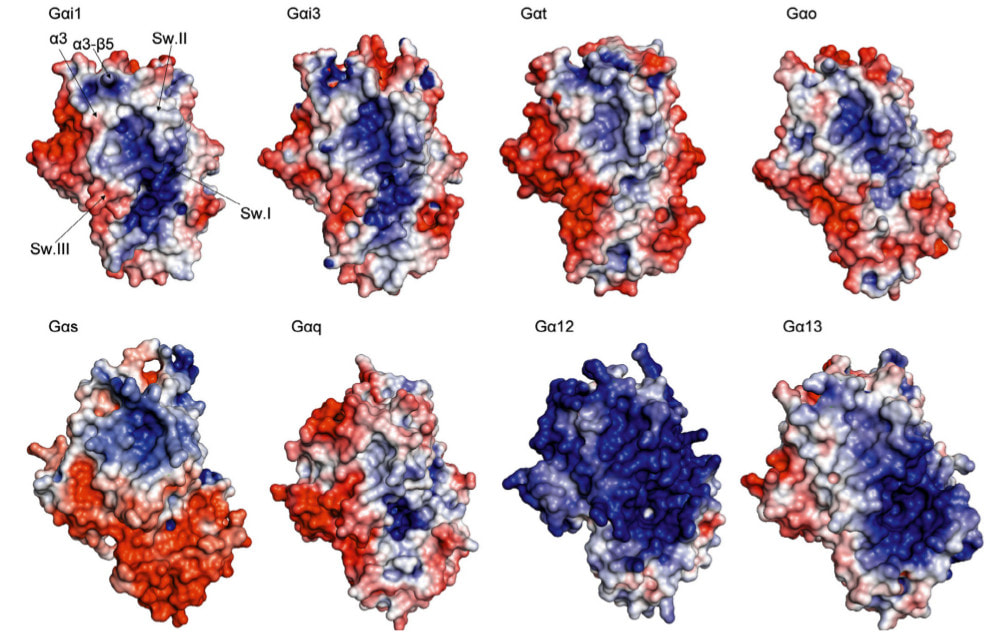
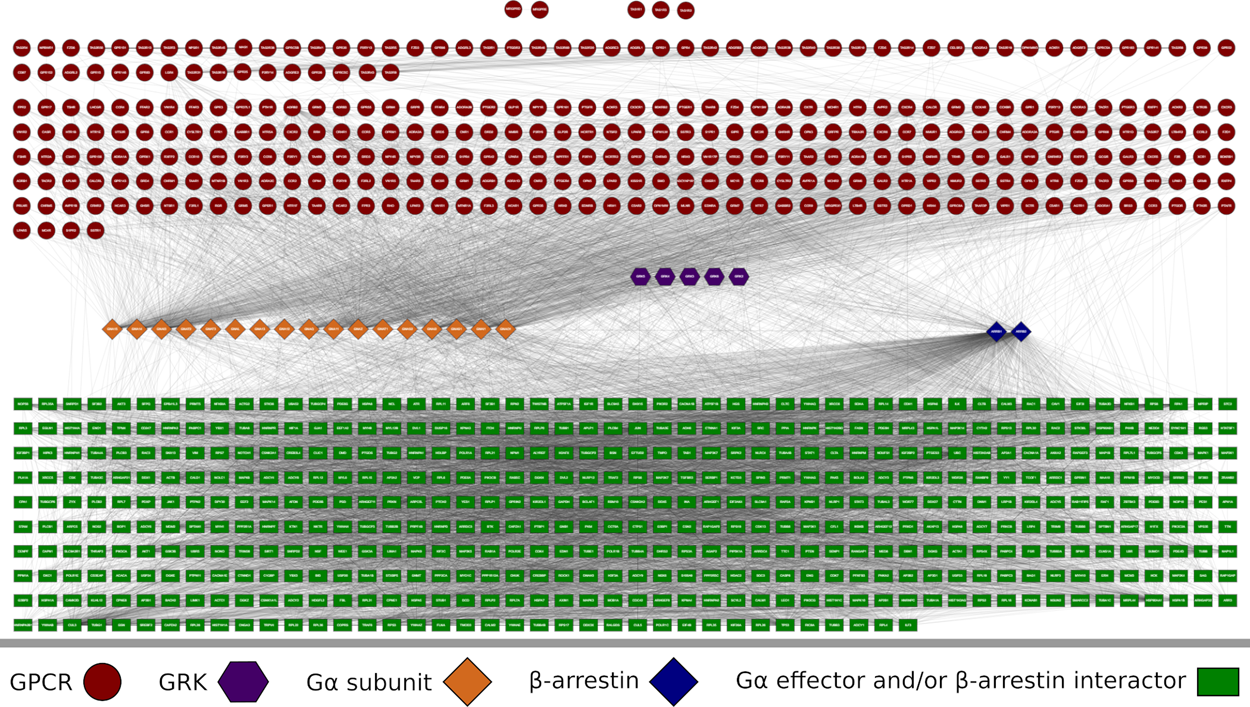
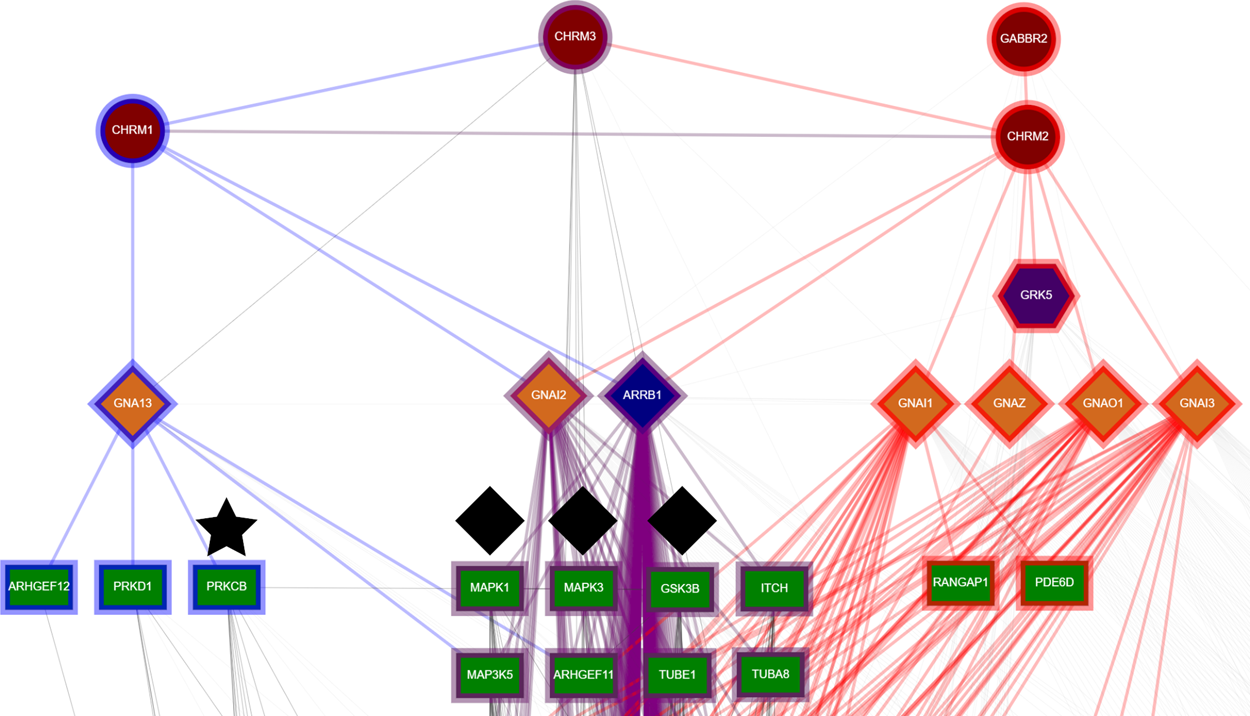
Media Gallery
Molecular Dynamics Simulations
Biological Networks
Journal Covers
Conference Posters
Conference Presentations
HeCrA 2018 |
Protein Summer School 2016 |
HSCBB 2015 |
HSCBB 2014 |
Videos
|
|
|